Why?
5.4 million people are bitten by snakes every year and 81-138 million people die due to snake bites each year. Preventing snake bites is clearly a major issue, in need of preventative measures to save lives. The project Snaked demonstrates a potential solution to alleviate the problem of struggling to identify the snake which has bitten a person. A proof of concept app is available on the Google Play Store which uses this model.
Challenge
- Each snake species varies in shape, colour, size, texture and more.
- Over 3000 snake species have already been discovered worldwide!
- Different snake species may look nearly identical, however, vary significantly
So it is clearly not an easy job to identify one snake from another, even though it's quintessential that we do.
Frameworks and Methods Utilized
PyTorch is used for ALL deep learning code and Numpy for numerical computations. The code is separated into the main file outlining the chosen algorithms (can be swapped/modified easily) and abstracted code to help train, evaluate and create an executable for any model easily.
There are several novel ideas/techniques used here which help to create neater/more readable pythonic code. The three primary examples of new PyTorch techniques:
- The Item tuple class which allows modular and further extensible code (when extra data needs to be processed)
- Use of super-convergence/the one-cycle policy in pure PyTorch instead of a highly abstracted library (i.e. Fast.AI)/bare python/numpy
- Use of a dictionary to dictate how different data sets should be split up to allow easy modifications of data proportions (i.e. switching between full dataset for training and small batches for ensuring all code runs without errors)
Although several models were trialed out on the dataset, in the end, a MobileNetV2 model provided the best results, whilst also remaining relatively lightweight and so able to run on low-power devices like phones (essential for the app). The final model was trained using LDAM loss instead of cross-entropy loss due to the dataset being imbalanced (some classes having far more samples than others). Note that the codebase has support for classical rebalancing, however, experimental trials show that this method causes overfitting extremely early on. The model manages to achieve around a 70% accuracy and F1 score. More details about the choice of model, how it was improved and lessons learnt from this project can be found on The Data Science Swiss Army Knife blog.
Android Application
The android application created for this project was written with Kotlin, using Fotoapparat (for easy camera support) and PyTorch (for utilizing the chosen PyTorch model) libraries. Due to the lightweight MobileNetV2 model no network is required to connect to a server (which would normally run the computations itself). This is intentionally done to facilitate use within remote locations! Please note that this is only a sample proof of concept app and if you're bitten consult a medical expert immediately.
Sources
All data currently used for the project comes from AIcrowd's Snake Species Identification Challenge. The images and labels have been used, however, geographic locations have been ignored to allow easy usage for any image, even if it hasn't been tagged. The dataset allows 85 species to be labelled. A Jupyter Notebook is also provided which demonstrates how to collect further data using Google Image searches! All statistics used here, or within the repository are from the World Health Organisation (unless otherwise stated) or pertaining specifically to the Snaked source code.
For further information please see the following:
- Snakebite envenoming
- Venomous Snakes Distribution and Species Risk Categories
- AIcrowd Snake Species Identification Challenge
Metrics
Throughout this report, I'll refer to F1 scores as my primary metric. This is because of the class imbalance. Please ensure to note that there is a major difference between the support number and number of present samples for a class. The first refers to the count in the validation dataset, whereas the latter in the training dataset. The number present in the training set will be primarily used to judge the effect of a skewed dataset on model predictions.
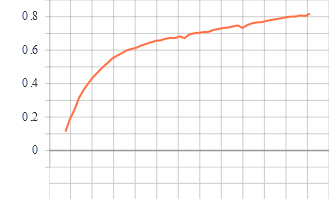
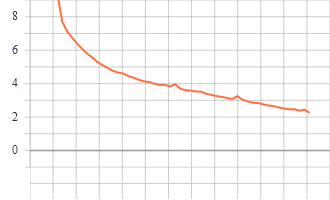
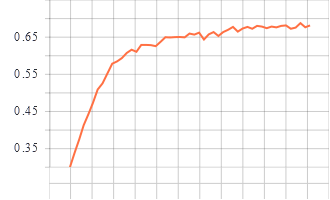
Training and Validation Graphs
Train epoch vs accuracy
Train epoch vs loss
Validation epoch vs accuracy
Validation epoch vs loss
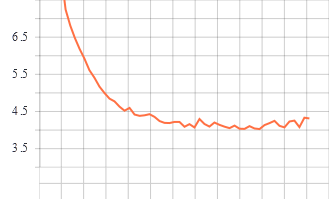
Model trained for 51 epochs
Classification Report
| species | precision | recall | f1-score | support |
| agkistrodon-contortrix | 0.8452380952380952 | 0.8765432098765432 | 0.8606060606060606 | 81.0 |
| agkistrodon-piscivorus | 0.676923076923077 | 0.6027397260273972 | 0.6376811594202899 | 73.0 |
| ahaetulla-prasina | 0.8 | 0.6666666666666666 | 0.7272727272727272 | 6.0 |
| arizona-elegans | 0.631578947368421 | 0.5714285714285714 | 0.6 | 21.0 |
| boa-imperator | 0.6 | 0.5294117647058824 | 0.5625 | 17.0 |
| bothriechis-schlegelii | 0.8888888888888888 | 0.7619047619047619 | 0.8205128205128205 | 21.0 |
| bothrops-asper | 0.875 | 0.5384615384615384 | 0.6666666666666667 | 13.0 |
| carphophis-amoenus | 0.6551724137931034 | 0.6551724137931034 | 0.6551724137931034 | 29.0 |
| charina-bottae | 0.8947368421052632 | 0.7391304347826086 | 0.8095238095238095 | 23.0 |
| coluber-constrictor | 0.5689655172413793 | 0.559322033898305 | 0.5641025641025641 | 59.0 |
| contia-tenuis | 0.52 | 0.6190476190476191 | 0.5652173913043478 | 21.0 |
| coronella-austriaca | 0.3333333333333333 | 0.1 | 0.15384615384615383 | 10.0 |
| crotalus-adamanteus | 0.6153846153846154 | 0.7272727272727273 | 0.6666666666666667 | 11.0 |
| crotalus-atrox | 0.7747252747252747 | 0.8392857142857143 | 0.8057142857142857 | 168.0 |
| crotalus-cerastes | 0.7857142857142857 | 0.8461538461538461 | 0.8148148148148148 | 13.0 |
| crotalus-horridus | 0.7741935483870968 | 0.8135593220338984 | 0.7933884297520662 | 59.0 |
| crotalus-molossus | 0.6363636363636364 | 0.7 | 0.6666666666666666 | 10.0 |
| crotalus-oreganus | 0.5333333333333333 | 0.6666666666666666 | 0.5925925925925926 | 12.0 |
| crotalus-ornatus | 1.0 | 0.8181818181818182 | 0.9 | 11.0 |
| crotalus-pyrrhus | 0.8571428571428571 | 0.5454545454545454 | 0.6666666666666665 | 22.0 |
| crotalus-ruber | 0.7894736842105263 | 0.7142857142857143 | 0.7500000000000001 | 21.0 |
| crotalus-scutulatus | 0.8620689655172413 | 0.7575757575757576 | 0.8064516129032258 | 33.0 |
| crotalus-viridis | 0.5384615384615384 | 0.6363636363636364 | 0.5833333333333334 | 22.0 |
| diadophis-punctatus | 0.8266666666666667 | 0.7948717948717948 | 0.8104575163398693 | 78.0 |
| epicrates-cenchria | 1.0 | 0.5 | 0.6666666666666666 | 2.0 |
| haldea-striatula | 0.5454545454545454 | 0.47058823529411764 | 0.5052631578947367 | 51.0 |
| heterodon-nasicus | 0.7 | 0.5384615384615384 | 0.608695652173913 | 13.0 |
| heterodon-platirhinos | 0.7058823529411765 | 0.75 | 0.7272727272727272 | 48.0 |
| hierophis-viridiflavus | 0.5 | 0.4375 | 0.4666666666666667 | 16.0 |
| hypsiglena-jani | 0.5238095238095238 | 0.6111111111111112 | 0.5641025641025642 | 18.0 |
| lampropeltis-californiae | 0.8701298701298701 | 0.8170731707317073 | 0.8427672955974842 | 82.0 |
| lampropeltis-getula | 0.8333333333333334 | 0.625 | 0.7142857142857143 | 16.0 |
| lampropeltis-holbrooki | 0.6923076923076923 | 0.6428571428571429 | 0.6666666666666666 | 14.0 |
| lampropeltis-triangulum | 0.8166666666666667 | 0.8305084745762712 | 0.8235294117647058 | 59.0 |
| lichanura-trivirgata | 0.9333333333333333 | 0.8235294117647058 | 0.8749999999999999 | 17.0 |
| masticophis-flagellum | 0.5294117647058824 | 0.6428571428571429 | 0.5806451612903226 | 42.0 |
| micrurus-tener | 1.0 | 0.95 | 0.9743589743589743 | 20.0 |
| morelia-spilota | 0.4 | 0.2222222222222222 | 0.2857142857142857 | 9.0 |
| naja-naja | 0.8333333333333334 | 0.7142857142857143 | 0.7692307692307692 | 7.0 |
| natrix-maura | 0.25 | 0.125 | 0.16666666666666666 | 8.0 |
| natrix-natrix | 0.7857142857142857 | 0.4074074074074074 | 0.5365853658536585 | 27.0 |
| natrix-tessellata | 0.5 | 0.36363636363636365 | 0.4210526315789474 | 11.0 |
| nerodia-cyclopion | 0.5833333333333334 | 0.4666666666666667 | 0.5185185185185186 | 15.0 |
| nerodia-erythrogaster | 0.53125 | 0.4358974358974359 | 0.47887323943661975 | 78.0 |
| nerodia-fasciata | 0.6190476190476191 | 0.43333333333333335 | 0.5098039215686274 | 30.0 |
| nerodia-rhombifer | 0.7213114754098361 | 0.6567164179104478 | 0.6875 | 67.0 |
| nerodia-sipedon | 0.5084745762711864 | 0.5309734513274337 | 0.5194805194805195 | 113.0 |
| nerodia-taxispilota | 0.8461538461538461 | 0.55 | 0.6666666666666667 | 20.0 |
| opheodrys-aestivus | 0.9042553191489362 | 0.9444444444444444 | 0.9239130434782609 | 90.0 |
| opheodrys-vernalis | 0.8235294117647058 | 0.6666666666666666 | 0.7368421052631577 | 21.0 |
| pantherophis-alleghaniensis | 0.36585365853658536 | 0.4 | 0.38216560509554137 | 75.0 |
| pantherophis-emoryi | 0.5714285714285714 | 0.6666666666666666 | 0.6153846153846153 | 36.0 |
| pantherophis-guttatus | 0.9183673469387755 | 0.8035714285714286 | 0.8571428571428571 | 56.0 |
| pantherophis-obsoletus | 0.5395348837209303 | 0.6041666666666666 | 0.5700245700245701 | 192.0 |
| pantherophis-spiloides | 0.43478260869565216 | 0.25 | 0.3174603174603175 | 40.0 |
| pantherophis-vulpinus | 0.5957446808510638 | 0.8484848484848485 | 0.7 | 33.0 |
| phyllorhynchus-decurtatus | 0.6956521739130435 | 0.9411764705882353 | 0.7999999999999999 | 17.0 |
| pituophis-catenifer | 0.7073170731707317 | 0.7016129032258065 | 0.7044534412955465 | 124.0 |
| pseudechis-porphyriacus | 0.6666666666666666 | 0.2857142857142857 | 0.4 | 7.0 |
| python-bivittatus | 0.875 | 0.7777777777777778 | 0.823529411764706 | 9.0 |
| python-regius | 0.0 | 0.0 | 0.0 | 3.0 |
| regina-septemvittata | 0.56 | 0.6666666666666666 | 0.6086956521739131 | 21.0 |
| rena-dulcis | 0.6666666666666666 | 0.8 | 0.7272727272727272 | 10.0 |
| rhinocheilus-lecontei | 0.8292682926829268 | 0.85 | 0.8395061728395061 | 40.0 |
| sistrurus-catenatus | 1.0 | 0.8 | 0.888888888888889 | 5.0 |
| sistrurus-miliarius | 1.0 | 0.6666666666666666 | 0.8 | 6.0 |
| sonora-semiannulata | 0.3333333333333333 | 0.4 | 0.3636363636363636 | 5.0 |
| storeria-dekayi | 0.76 | 0.8465346534653465 | 0.8009367681498829 | 202.0 |
| storeria-occipitomaculata | 0.631578947368421 | 0.6486486486486487 | 0.64 | 37.0 |
| tantilla-gracilis | 0.5454545454545454 | 0.6 | 0.5714285714285713 | 10.0 |
| thamnophis-cyrtopsis | 0.5714285714285714 | 0.4444444444444444 | 0.5 | 9.0 |
| thamnophis-elegans | 0.47058823529411764 | 0.2962962962962963 | 0.3636363636363636 | 27.0 |
| thamnophis-hammondii | 0.5555555555555556 | 0.35714285714285715 | 0.43478260869565216 | 14.0 |
| thamnophis-marcianus | 0.8787878787878788 | 0.8285714285714286 | 0.8529411764705883 | 35.0 |
| thamnophis-ordinoides | 0.375 | 0.16666666666666666 | 0.23076923076923078 | 18.0 |
| thamnophis-proximus | 0.7543859649122807 | 0.7413793103448276 | 0.7478260869565219 | 58.0 |
| thamnophis-radix | 0.8181818181818182 | 0.47368421052631576 | 0.6 | 38.0 |
| thamnophis-sirtalis | 0.7167070217917676 | 0.896969696969697 | 0.7967698519515478 | 330.0 |
| tropidoclonion-lineatum | 0.7777777777777778 | 0.6363636363636364 | 0.7000000000000001 | 11.0 |
| vermicella-annulata | 0.0 | 0.0 | 0.0 | 3.0 |
| vipera-aspis | 0.4 | 0.5714285714285714 | 0.47058823529411764 | 7.0 |
| vipera-berus | 0.75 | 0.75 | 0.75 | 12.0 |
| virginia-valeriae | 0.2 | 0.14285714285714285 | 0.16666666666666666 | 7.0 |
| xenodon-rabdocephalus | 1.0 | 1.0 | 1.0 | 8.0 |
| zamenis-longissimus | 0.4 | 0.3333333333333333 | 0.3636363636363636 | 12.0 |
| accuracy | 0.6930662557781202 | 0.6930662557781202 | 0.6930662557781202 | 0.6930662557781202 |
| macro avg | 0.6659430597272402 | 0.6050948460385794 | 0.6247619446039016 | 3245.0 |
| weighted avg | 0.6925634979734435 | 0.6930662557781202 | 0.6866874083865039 | 3245.0 |
Despite a skewed dataset, the majority of snakes had a similar precision and recall score.
Number of Samples vs F1 score
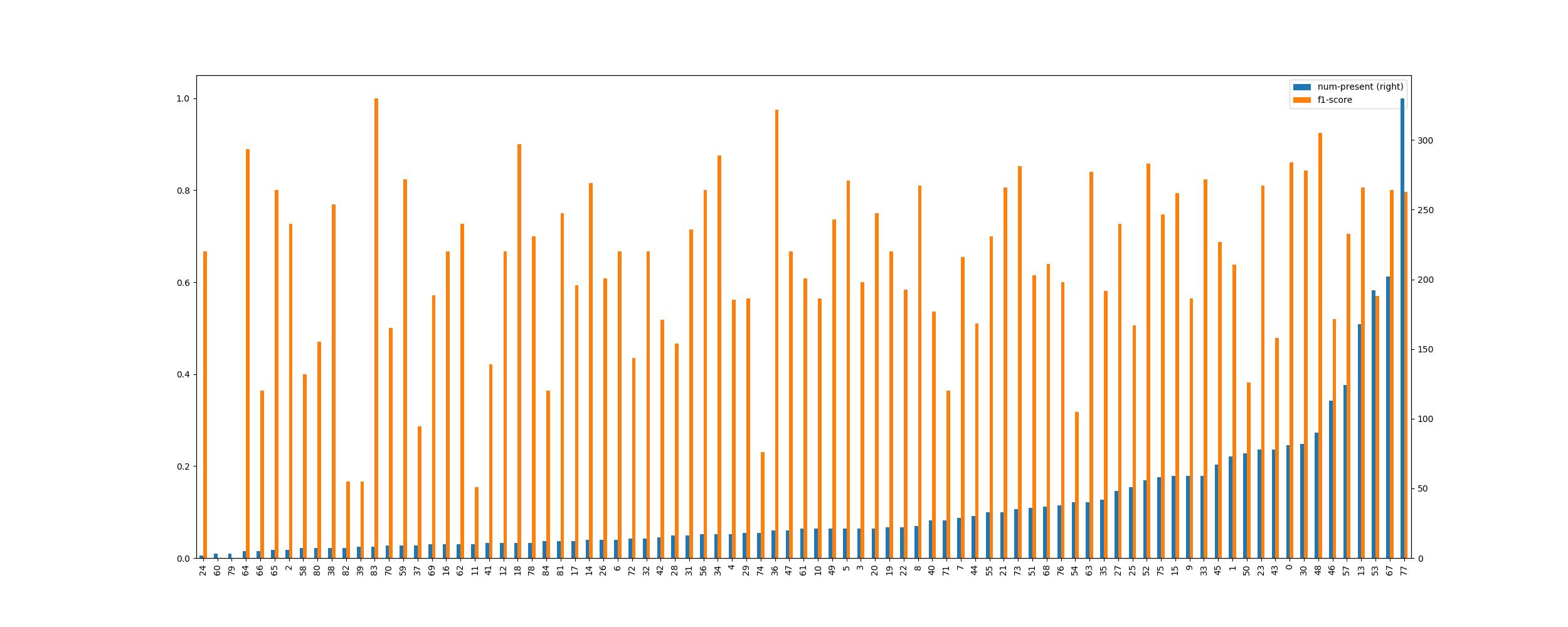 The above graph of the number of samples the neural network was trained over and F1 score per class indicates two significant points:
The above graph of the number of samples the neural network was trained over and F1 score per class indicates two significant points:
- Having more samples of a class will increase the probability of a model robustly identifying the snake species correctly
- However, this does not mean that an inability to collect a large number of images for all classes will create a major imbalance in a model's predictions
The latter takeaway shows that LDAM loss has been largely successful! On a side note though, when a class has few samples, the F1 score may not be completely representative of how the model will generalise. This is primarily because, in a small number of images, only a small number of conditions can be shown and tested. Yet, in reality, an image can be taken in any environment and transformed in a very large number of ways.
Confusion Matrix
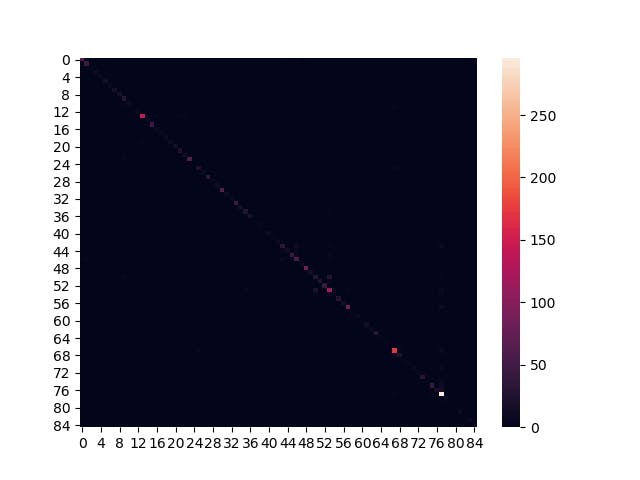
The test dataset had too few images to clearly judge anything from the confusion matrix. This is due to a combination of having 85 classes, and classes with few samples being quite dark.
Improvements
If this model was going to be retrained, a good idea would be to use a 90-5-5, or better a 80-10-10 split. This would ensure that there would be enough data in the testing dataset to judge where exactly the model's confusion stems from (using a confusion matrix).
Combining the current classification model with an additional preprocessing pipeline which includes segmentation may also allow far higher F1 scores. This is because segmentation is able to remove extraneous noise around a snake (subtle environmental cues which prevent the model from generalising). Additionally, this would completely avoid the chance of a snake being cropped out of an image (currently unlikely but still possible for small snakes).